Oxymatrine Inhibition of Hepatitis B Virus Replication Through ERK1/2 Pathway and HNF1α and HNF4α Block in vitro
By Fangsheng Wang, Xiaoling Wang, Baodian Ling, Hongfei Lu, Junyun Huang, Tianyu ZhongAffiliations
doi: 10.29271/jcpsp.2024.03.329ABSTRACT
Objective: To explore the molecular mechanism of oxymatrine (OM) by increasing the phosphorylation of ERK1/2 signal factor and blocking the transcription factors HNF1α and HNF4α expression against hepatitis B virus (HBV) antigen secretion and HBV DNA replication in HepG2.2.15 cells.
Study Design: An experimental study.
Place and Duration of the Study: Department of Laboratory Medicine, First Affiliated Hospital of Gannan Medical University, Jiangxi, China, between May 2020 and December 2022.
Methodology: HepG2.2.15 cells, known for stably expressing HBV particles, were utilised as a cell-based model to explore potential pathways pertaining to the OM inhibition of HBV replication. An MTT assay was utilised to measure cytotoxicity. HBsAg or HBeAg content was measured using an enzyme-linked immunosorbent assay kit. HBV DNA in cell-free culture media was examined using a fluorescent quantitative PCR kit. Real-time PCR was utilised to analyse HNF1α and HNF4α mRNA expression, whereas Western blotting was performed to evaluate HNF1α, HNF4α, and ERK1/2 protein expression.
Results: OM inhibited HBV DNA copy number in the cell supernatant, 3.5-kb RNA gene expression in cells, and HBsAg and HBeAg secretion. OM upregulated p-ERK1/2 protein and significantly downregulated HNF1α and HNF4α gene transcription and protein translation.
Conclusion: OM may inhibit the replication of HBV by inducing the phosphorylation of ERK1/2 and blocking the transcription factors HNF1α and HNF4α expression that are essential for viral replication.
Key Words: Oxymatrine, ERK1/2, Hepatocyte nuclear factor, Anti-HBV.
INTRODUCTION
According to recent surveys and studies, the global infection count for Hepatitis B Virus (HBV) is 364 million individuals.1 Approximately 1 million individuals die each year from liver disease complications occasioned by HBV infection, and approximately 300,000 of them reside in China.2 Therefore, it is necessary to strengthen the early control of viral replication to effectively reduce the occurrence of liver complications.
Hepatocyte nuclear factors (HNFs) are transcription factors that are instrumental in regulating liver-specific genes expression and exert pivotal roles in liver differentiation and metabolism.3 HNF4α, in particular, functions as a dominant regulator of HBV gene replication and expression as well as a crucial regulator of the HBV liver-specific function.4
Thus, HNFs and HBV exert a vital regulatory impact on the persistent survival of HBV in the host.5 However, activating the ERK1/2, which are members of the mitogen-activated protein kinase (MAPK) family, signalling pathway can disrupt the formation of the HNF4α enhancer–promoter complex and affect HBV replication in human hepatoma cells.6 Oxymatrine (OM), a matrine-type alkaloid easily derived from the Chinese herb Sophora flavescent, exhibits a diverse range of pharmacological activities, such as anti-virus, anti-inflammatory, anti-fibrosis, and immune regulation.7,8 As previously reported, OM can directly inhibit HBV replication in vitro and in vivo, and is extensively utilised in the clinical therapy of viral hepatitis B;9,10 however, the anti-HBV mechanism of OM has not been fully elucidated.11,12 In preliminary real-time quantitative PCR experiments, OM was found to significantly downregulate HNF4α and HNF1α mRNA in HepG2.2.15 cells. This study aimed to apply HepG2.2.15 cells, production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA, as a cell model for observing and analysing the potential mechanistic pathways of OM in suppressing HBV replication.
METHODOLOGY
The study was conducted at the First Affiliated Hospital of Gannan Medical University, Jiangxi, China, between May 2020 and December 2022 after obtaining the approval from the Clinical Research Ethics Review Committee of the hospital. HepG2.2.15 cells were revived with a complete medium containing 10% newborn foetal bovine serum and Dulbecco's Modified Eagle Medium following the manufacturer's instructions and underwent a culture process at 37°C in a 5% CO2 incubator.
To investigate OM inhibition of HBV expression in HepG2.2.15 cells, the cells were classified into five groups according to the OM concentration as control (0 μg/mL), 10 μg/mL, 100 μg/mL, and 1000 μg/mL OM groups, and the 3.75 nM entecavir (ETV) (MCE, USA) group.13 In a 24-well plates setup, four duplicate wells were set in each group, with each well hosting 1 × 10.4 The cells underwent a 12-hour incubation period in a serum-free DMEM basal medium at 37°C in a 5% CO2 incubator. Groups had corresponding amount of OM or ETV in fresh media added, and were subsequently subjected to incubation at 37 °C and 5% CO2 lasting 3, 6, and 9 days to observe the cell state.
To investigate the mechanism of OM inhibiting HBV expression in HepG2.2.15 cells, the cells were classified into three groups: Control, 100 μg/mL, and 1000 μg/mL OM. Three duplicate wells were set for each group, the number of cells per well was 1 × 105/mL, and the corresponding drugs were added and incubated at 37°C and 5% CO2 lasting 24 or 48 h.
For assays on OM-induced ERK1/2 signalling after ERK1/2 inhibitor intervention, HepG2.2.15 cells were classified into three groups: DMSO, OM (100 μg/mL), and U0126(10 μM) (Beyotime, China) +OM (100 μg/mL). The U0126+OM group was pretreated with U0126—an ERK1/2 inhibitor—lasting 1 h. After discarding the supernatant and counting it as 0 h, OM at 100 μg/mL was added, which was subjected to further incubation with the cells at 37°C and 5% CO2 lasting 4, 12, and 24 h.
After culturing the cells of the Control, 10, 100, and 1000 μg/mL OM groups for 9 days, MTT (5 mg/mL) was added. A 4-hour incubation period was followed by measurement of the absorbance value of the wells at 490 nm, which was detected using a microplate reader, and the cell viability percentage was determined:
Cell survival rate = (Experimental group − Blank group) / (Control group − Blank group).
An ELISA kit (Kehua, Shanghai, China) was utilised to measure HBsAg and HBeAg levels in cell culture supernatants at 3, 6, and 9 days or at 24 and 48 h after the appropriate interventions. The ELISA was conducted strictly following the manufacturer’s instructions.
The cell supernatant and standard HBV DNA genome extraction strictly followed the instructions contained in the DNA virus extraction kit, and the HBV DNA copies in the cell supernatant were determined at 3, 6, and 9 days (primer sequences provided in Table I). The extraction of intracellular total RNA and the reverse transcription process for synthesising the first strand of cDNA were also performed following the reverse transcription kit instructions (Kangwei Century, China). GAPDH was utilised as the internal reference gene for RT-qPCR (Bio-Rad, USA). HNF1α, HNF4α, and 3.5-kb RNA genes expression in 48-h cells were analysed by the 2−ΔΔCt method (primer sequences provided in Table I). Amplification conditions for the qPCR reaction system (50 μL) were pre-denaturation at 95 °C lasting 10 min; 40 cycles of denaturation at 95°C lasting 15 s and annealing at 60°C lasting 1 min; and melting curve analysis at 95 °C lasting 5 s and at 65°C lasting 5 s. The standard curve was established based on the Ct value of the standard substance, the regression equation was calculated, and the HBV DNA copy number in the sample was determined according to the regression equation. The HBV DNA inhibition rate was subsequently determined using the following formula:
Inhibitory rate = (Control − Experimental) / Control × 100%
Western blotting was utilised to measure HNF1α, HNF4α, and p-ERK1/2 proteins expression in cells after 9 days of culture as well as HNF1α, HNF4α, and p-ERK1/2 in cells at 0, 4, 12, and 24 h. Total intracellular protein was extracted following the reagent instructions and lysed using 150 μL of RIPA. All lysed samples were collected, and the protein level of each cell lysate was accessed by a BCA protein assay kit (Beyotime, China). Subsequently, 10 μL of protein per well was electrophoresed on a 5%–12% concentration and separating gel, which was subsequently placed on a PVDF membrane. The membrane was subjected to a blocking step with 5% non-fat milk powder lasting 1.5 h and, subsequently, incubated with a 1:1000 dilution of primary antibody (Boao Biological, China) at room temperature lasting 2 h with shaking at low speed, followed by overnight incubation at 4°C. After washing to remove the primary antibody, the membrane was subjected to a horseradish peroxidase-conjugated secondary antibody, utilised at a 1:7000 dilutions, and incubated for 1.5 h at room temperature on a low-speed shaker. The exposure was performed with an ultra-sensitive ECL chemiluminescence kit, and the bands were analysed by the grey value of Image J software.
The statistical software SPSS 27.0 was utilised for analysis, and the experimental data were represented as mean ± standard deviation. Comparisons between groups were analysed by the one-way analysis of variance (ANOVA) method. When the variance was equal, the least significant difference (LSD) method was utilised for multiple comparisons between groups. p <0.05 was deemed statistically significant.
Table I: qPCR primer sequences for HBV DNA and for HNF1α, HNF4α, and 3.5-kb RNA.
|
Primer name |
Primer sequence (5′→3′) |
|
HBV DNA |
F: TGTCCTGGTTATCGCTGG |
|
R: CAAACGGGCAACATACCTT |
|
|
3.5-kb RNA |
F: CTCAATCTCGGGAATCTCAATGT |
|
R: TGGATAAAACCTAGCAGGCATAAT |
|
|
HNF1α |
F: TCTACAACTGGTTTGCCAACC |
|
R: GGCTTCTGTACTCAGCAGGC |
|
|
HNF4α |
F: ACGGGCAAACACTACGG |
|
R: ATTCTGGACGGCTTCCTT |
|
|
GAPDH |
F: ACAGCCTCAAGATCATCAGCA |
|
R: ATGAGTCCTTCCACGATACCA |
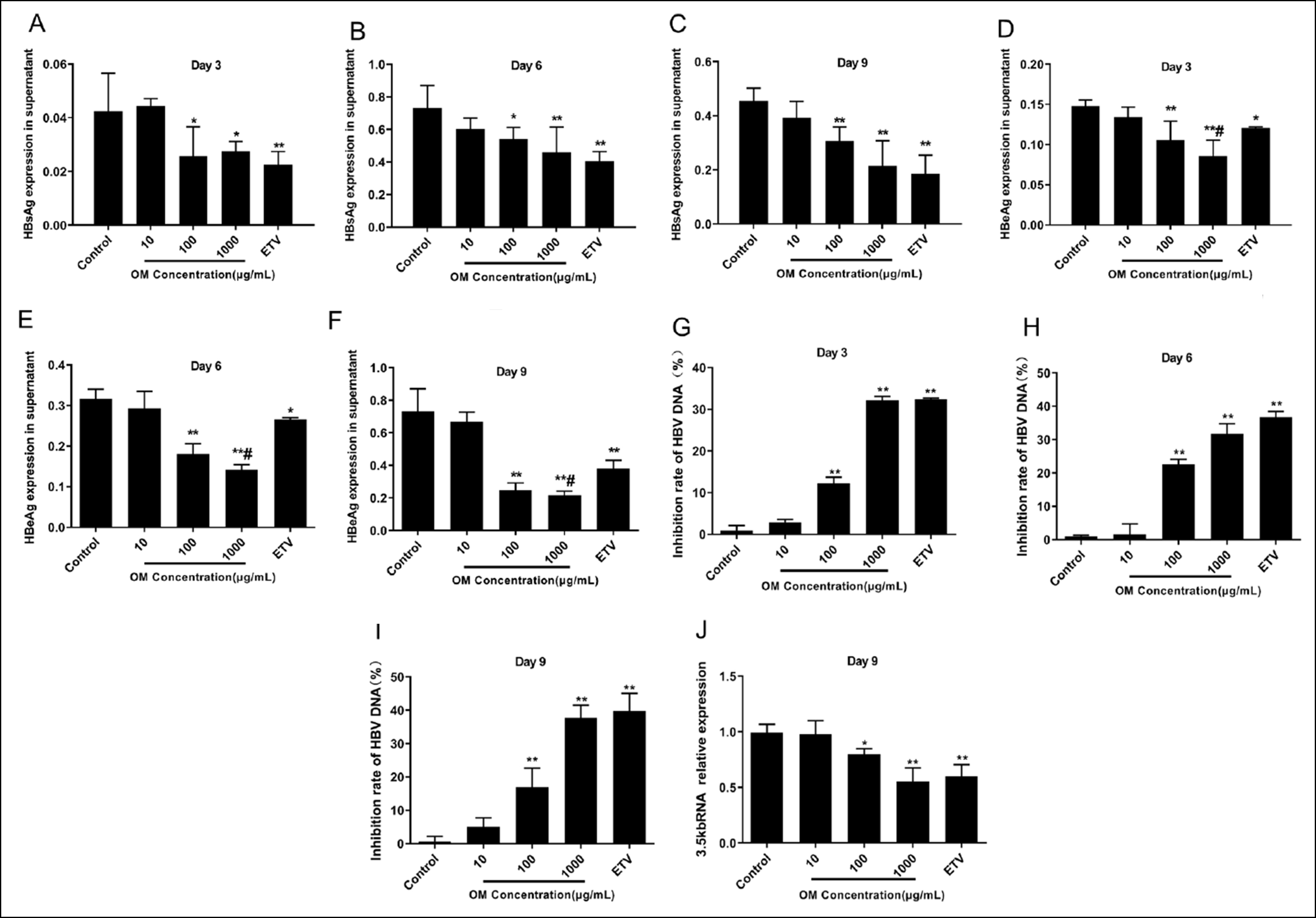 Figure 1: OM inhibits the level of HBsAg, HBeAg, HBV DNA and intracellular 3.5-kb RNA. A-F: OM suppresses HBsAg and HBeAg secretion from HepG2.2.15 cells. G-J: OM inhibits the expression levels of supernatant HBV DNA and intracellular 3.5-kb RNA in HepG2.2.15 cells.
Figure 1: OM inhibits the level of HBsAg, HBeAg, HBV DNA and intracellular 3.5-kb RNA. A-F: OM suppresses HBsAg and HBeAg secretion from HepG2.2.15 cells. G-J: OM inhibits the expression levels of supernatant HBV DNA and intracellular 3.5-kb RNA in HepG2.2.15 cells.** p < 0.01, * p < 0.05 compared to Control and # p < 0.05 compared to ETV.
RESULTS
After the HepG2.2.15 cells were subjected to OM lasting 9 days, the cell viability (n = 3, 95.67 ± 0.53%) of 1000μg/mL groups remained above 95%. Thus, the cytotoxicity of OM was low.
Inhibitory impact of OM on HBsAg and HBeAg. Post incubation of HepG2.2.15 cells with diverse concentrations of OM lasting 3, 6, and 9 days was performed, and HBsAg and HBeAg levels in the cell culture supernatants were measured (Figure 1A-F). In comparison to the Control group, a notable decline in HBsAg and HBeAg was observed in the supernatant of the ETV group (p < 0.01 or p < 0.05), indicating an impaired secretion of these proteins. OM at 100 and 1000 μg/mL also greatly suppressed HBsAg and HBeAg secretion in cells (p < 0.01 or p < 0.05). As OM concentration escalated, a gradually decline in HBeAg secretion was observed, indicating the existence of a dose-dependent effect (p < 0.01 or p < 0.05). A prominently reduced level of HBeAg in the culture supernatant after 1000μg/mL OM treatment was noted compared with that of ETV treatment (Figure 1E and F) (p < 0.01 or p <0.05). These results indicated that OM treatment led to a dose-dependent suppression of HBsAg and HBeAg secretion in HepG2.2.15 cells.
OM inhibits the level of HBV DNA and intracellular 3.5-kb RNA. Following 3, 6, and 9 days of culture (Figure 1G–I), contrary to the Control group, the ETV group demonstrated a substantial inhibitory effect on HBV DNA (p < 0.01), and the HepG2.2.15 cells processed with 100 and 1000 μg/mL OM also exhibited a notable increase in the inhibition rate of HBV DNA in the culture supernatant. Following 9 days of culture, in comparison to the Control group, the 3.5-kb RNA in the cells treated with 100 and 1000 μg/mL OM was considerably reduced (p < 0.01, Figure 1J). Furthermore, with increasing OM concentrations, the inhibition of HBV DNA and 3.5-kb RNA was gradually enhanced (p < 0.01).
HNF1α, HNF4α mRNA, and 3.5-kb RNA expression. After culture for 24 and 48 h, compared with Control, HBsAg and HBeAg secretion in the cell supernatant of the 100 and 1000 μg/ml OM declined considerably (p < 0.01, Figure 2A and B). Concurrently, HNF1α and HNF4α mRNA and 3.5-kb RNA in the HepG2.2.15 cells subjected to 100 and 1000 μg/mL OM were notably downregulated (p < 0.01 or p < 0.05, Figure 2C-E). The inhibitory effect was more significant with increasing OM concentration. These results indicated that OM suppressed the gene expression of HNF1α and HNF4α and 3.5-kb RNA in vitro.
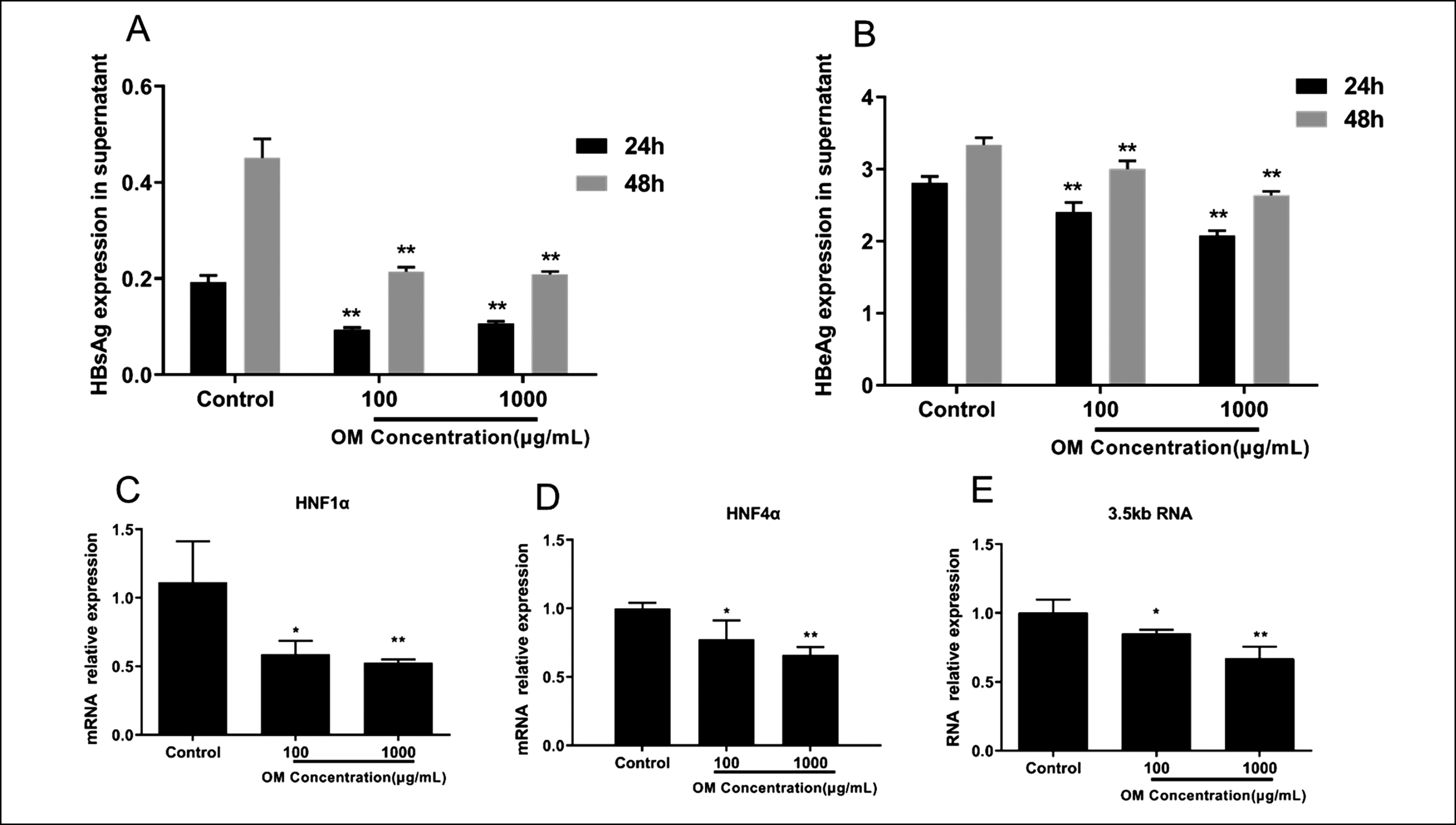 Figure 2: Relative expression levels of 3.5-kb RNA, HNF1α, and HNF4α mRNA in HepG2.2.15 cells after OM treatment. A and B: Comparison of HBsAg and HBeAg levels in the supernatant of each group after 24 and 48 h. C–E: Comparison of the relative expression of HNF1α mRNA (C), HNF4α mRNA (D), and 3.5-kb RNA (E) in each group after 48 h. ** p < 0.01, and * p < 0.05 compared to the Control.
Figure 2: Relative expression levels of 3.5-kb RNA, HNF1α, and HNF4α mRNA in HepG2.2.15 cells after OM treatment. A and B: Comparison of HBsAg and HBeAg levels in the supernatant of each group after 24 and 48 h. C–E: Comparison of the relative expression of HNF1α mRNA (C), HNF4α mRNA (D), and 3.5-kb RNA (E) in each group after 48 h. ** p < 0.01, and * p < 0.05 compared to the Control.
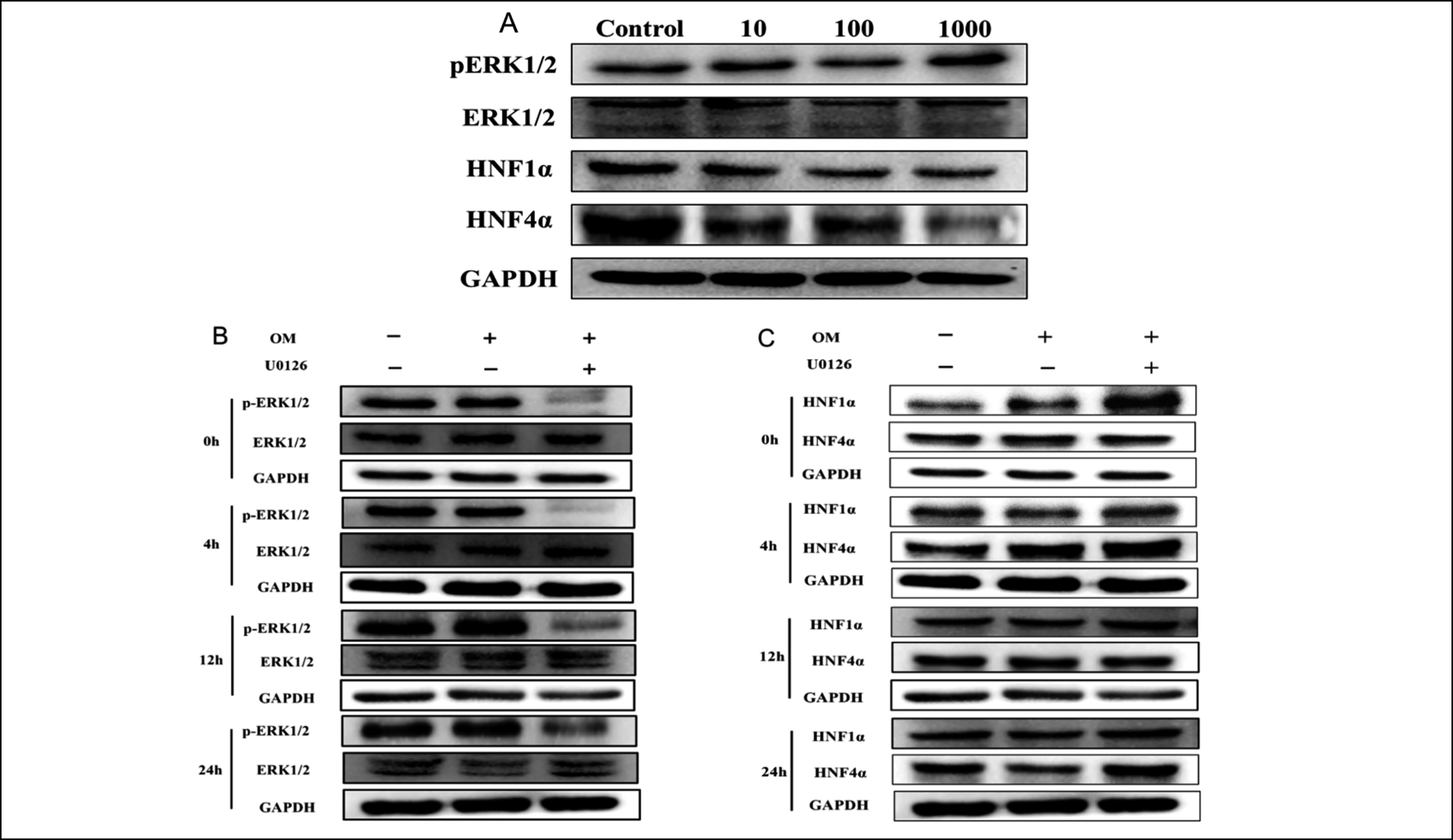 Figure 3: p-ERK1/2, HNF1α, and HNF4α levels in HepG2.2.15 cells post OM treatment. A: Relative expression of p-ERK1/2, HNF1α, and HNF4α proteins in HepG2.2.15 cells processed with various concentrations of OM for 9 days. B and C: p-ERK1/2, HNF1α, and HNF4α levels in HepG2.2.15 cells at various time points post-OM treatment. p-ERK1/2(B) and HNF1α and HNF4α(C) were detected by western blotting at 0–24 h.
Figure 3: p-ERK1/2, HNF1α, and HNF4α levels in HepG2.2.15 cells post OM treatment. A: Relative expression of p-ERK1/2, HNF1α, and HNF4α proteins in HepG2.2.15 cells processed with various concentrations of OM for 9 days. B and C: p-ERK1/2, HNF1α, and HNF4α levels in HepG2.2.15 cells at various time points post-OM treatment. p-ERK1/2(B) and HNF1α and HNF4α(C) were detected by western blotting at 0–24 h.
HNF1α, HNF4α, and p-ERK1/2 proteins expression. When compared with the control group, following 9 days of culture, the p-ERK1/2 level was greatly upregulated in the 100 and 1000 μg/mL OM-treated groups (p <0.01 or p < 0.05) (Figure 3A and Table II). Simultaneously, the protein expression of HNF1α and HNF4α was remarkably downregulated (p < 0.01). These results indicated that OM upregulated the expression of ERK1/2 and downregulated HNF1α and HNF4α expression in vitro.
Table II: p-ERK1/2, HNF1α, and HNF4α levels in HepG2.2.15 cells post OM treatment.
|
Proteins |
Time |
Groups |
Mean ± Standard |
p-value |
|
p-ERK1/2 |
9d |
Control |
0.54 ± 0.15 |
|
|
p-ERK1/2 |
9d |
10 |
0.49 ± 0.30 |
|
|
p-ERK1/2 |
9d |
100 |
1.33 ± 0.25 |
<0.01** |
|
p-ERK1/2 |
9d |
1000 |
1.64 ± 0.18 |
<0.01** |
|
HNF1α |
9d |
Control |
0.36 ± 0.05 |
|
|
HNF1α |
9d |
10 |
0.31 ± 0.05 |
|
|
HNF1α |
9d |
100 |
0.20 ± 0.03 |
<0.01** |
|
HNF1α |
9d |
1000 |
0.18 ± 0.03 |
<0.01** |
|
HNF4α |
9d |
Control |
0.36 ± 0.05 |
|
|
HNF4α |
9d |
10 |
0.33 ± 0.04 |
|
|
HNF4α |
9d |
100 |
0.20 ± 0.03 |
<0.01** |
|
HNF4α |
9d |
1000 |
0.16 ± 0.01 |
<0.01** |
|
p-ERK1/2 |
0h |
DMSO |
0.49 ± 0.12 |
|
|
p-ERK1/2 |
0h |
OM |
0.55 ± 0.14 |
|
|
p-ERK1/2 |
0h |
U0126+OM |
0.04 ± 0.01 |
<0.01** |
|
p-ERK1/2 |
24h |
DMSO |
0.34 ± 0.02 |
|
|
p-ERK1/2 |
24h |
OM |
0.46 ± 0.02 |
0.022* |
|
p-ERK1/2 |
24h |
U0126+OM |
0.16 ± 0.01 |
|
|
HNF1α |
0h |
DMSO |
0.31 ± 0.08 |
|
|
HNF1α |
0h |
OM |
0.35 ± 0.05 |
|
|
HNF1α |
0h |
U0126+OM |
0.48 ± 0.07 |
0.024* |
|
HNF1α |
24h |
DMSO |
0.25 ± 0.03 |
|
|
HNF1α |
24h |
OM |
0.16 ± 0.01 |
0.040* |
|
HNF1α |
24h |
U0126+OM |
0.28±0.06 |
|
|
HNF4α |
0h |
DMSO |
0.91 ± 0.14 |
|
|
HNF4α |
0h |
OM |
0.89 ± 0.17 |
|
|
HNF4α |
0h |
U0126+OM |
1.09 ± 0.15 |
0.028* |
|
HNF4α |
24h |
DMSO |
0.37 ± 0.04 |
|
|
HNF4α |
24h |
OM |
0.26 ± 0.04 |
0.018* |
|
HNF4α |
24h |
U0126+OM |
0.35 ± 0.06 |
|
|
Data are shown as relative protein expression. **p <0.01 and *p <0.05 compared to Control or DMSO. |
||||
The effect of OM on the ERK1/2 signalling pathway after the intervention of the ERK1/2 inhibitor U0126. Compared with the DMSO-treated group, HepG2.2.15 cells were pretreated with U0126 (10 μM) lasting 1 h followed by OM (100 μg/mL) lasting 24 h, the ERK1/2 phosphorylation triggered by OM was suppressed (p < 0.01, Figure 3B and Table II), and the HNF1α and HNF4α levels were greatly upregulated (p < 0.01, Figure 3C and Table II). These results verified that the inhibition of ERK1/2 phosphorylation could promote the expression of HNF1α and HNF4α. At 24 h, compared with the DMSO-treated group, p-ERK1/2 expression in the OM-treated cells was markedly upregulated (p < 0.05), and HNF1α and HNF4α expression was remarkably downregulated (p < 0.05). These findings further revealed that OM could suppress HBV replication by triggering the ERK1/2 pathway and downregulating HNF4α and HNF1α in HepG2.2.15 cells.
DISCUSSION
OM, which is an alkaloid derived from the Chinese herb Sophora flavescens, can inhibit HBV replication. Although its efficacy is similar to that of interferon-α (IFN-α), the mechanism of action remains unclear.14,15 OM, as a strong immunomodulator, activates the Toll-like receptor 9 (TLR9) signal transduction function, induces peripheral lymphocytes to secrete antiviral cytokines, cooperates with TLR9 ligands to boost the immune functionality of chronic hepatitis B patients, and inhibits HBV replication in vivo.16 Furthermore, OM accelerated the production of interferon-γ (Interferon-α, IFN-γ) in CD4+ T cells in a dose-responsive manner, enhanced the immune function of mice, and outperformed Entecavir in eliminating serum HBsAg and intrahepatic HBcAg.11
HNFs include the HNF1, HNF3, HNF4, and HNF6 families, with the HNF1 family having two subtypes (HNF1α and HNF1β) and the HNF4 family having three subtypes (HNF4α, HNF4β, and HNF4γ). HNF1α and HNF4α are responsible for HBV gene replication and are crucial regulators of antigen expression. HNF1α enhances viral transcription by activating the binding of HBV pre-S1P to its enhancer/promoter and the binding of HBV CP protein to HBV enhancer II. Overexpression of HNF4α upregulates the activities of pre-S1 and pre-S2 proteins and triggers HBV enhancer II, CP, and pre-S2P, thereby promoting HBV replication.17-20
In hepatoma cells within humans, the activation of MAPK signalling downregulates HNF4α expression and completely inhibits C/EBPα protein expression, and disrupts the recruitment of HNF3β, HNF1α, and RNA polymerase II to the HNF4α enhancer, thereby inhibiting 3.5-kb RNA synthesis and transcription.21,22 Zheng et al. unveiled that triggering the RAS-MAPK signalling pathway by external stimuli inhibited HBV replication in Huh7 and HepG2 cells, indicating that the MAPK signalling pathway exerts an inhibitory impact on some liver transcription factors.23
This study utilised HepG2.2.15 cells as a model to verify that OM suppressed HBV DNA, HBsAg, and HBeAg levels in a dose- and time-dependent manner. Moreover, OM was observed to inhibit the production of HBV 3.5-kb RNA. It was found that OM could increase the phosphorylation of ERK1/2 signalling factors, downregulate HNF1α and HNF4α, and affect 3.5-kb RNA expression. To further explore the mechanism of action of OM, U0126 was utilised to inhibit the phosphorylation of ERK1/2, and OM continued to stimulate the phosphorylation of ERK1/2 for different time periods. The expression levels of HNF4α and HNF4α were gradually downregulated, which proved that OM may block the protein expression of the transcription factors HNF1α and HNF4α that are required for virus replication by increasing the phosphorylation of ERK1/2.
Collectively, the activation of the MAPK signalling pathway exerts a vital role in anti-HBV activity. The stimulation of MAPK by external factors can result in the phosphorylation of signalling proteins to induce the transcription of various downstream genes with regulatory functions, including transcription factors, growth factors, kinases, and other enzymes.24,25 OM may increase the phosphorylation of ERK1/2, block the transcription factors HNF1α and HNF4 expression, which are essential for viral replication, and inhibit the synthesis of 3.5-kb RNA. This study demonstrated that OM (with U0126) induce the phosphorylation of ERK1/2 declined remarkably, and that HBsAg, HBeAg, and 3.5-kb RNA expression of HBV still exerts inhibitory effects, indicating that the effect of OM on HBV inhibition may be related to other mechanisms. Therefore, to obtain a more comprehensive understanding of the anti-HBV effect of OM and its molecular mechanism, future experiments can explore the JNK and P38 signalling factors in the MAPK signalling pathway. Such an understanding can lay an experimental foundation for detailed research on the application of OM in immune regulation.
CONCLUSION
Collectively, the results obtained from this study demonstrate that OM inhibits HBV DNA and 3.5kb-RNA as well as antigen secretion by upregulating the phosphorylation of ERK1/2 and by blocking the transcription factors HNF1α and HNF4α expression that are fundamental for viral replication.
FUNDING:
This work was supported by the Research Program of the Province Health Commission of Jiangxi (Grant/Award Number: 202310748) and the Research Program of the Municipal Health Commission of Ganzhou (Grant/Award Number: 2022-1-03).
ETHICAL APPROVAL:
This research project has been reviewed and approved to be appropriate and humane by the Clinical Research Ethics Review Committee of First Affiliated Hospital of Gannan Medical University, Ganzhou, China.
PATIENTS’ CONSENT:
Not applicable.
COMPETING INTEREST:
The authors declared no conflict of interest.
AUTHORS’ CONTRIBUTION:
FW, XW: Both authors contributed equally to conception and drafting of the manuscript.
JH, TZ: Critical intellectual input, contributed reagents, materials, and analysis tools.
BL: Statistical analysis of data.
HL: Performed the experiments.
All authors approved the final version of the manuscript to be published.
ACKNOWLEDGEMENTS:
The authors acknowledged the Precision Medicine Centre of First Affiliated Hospital of Gannan Medical University for providing an experimental platform.
REFERENCES
- OC P. Global prevalence, treatment, and prevention of hepatitis b virus infection in 2016: A Modelling Study. Lancet Gastroenterol Hepatol 2018; 3(6):383-403. doi: 10.1016/s2468-1253(18)30056-6.
- Xu R, Hu P, Li Y, Tian A, Li J, Zhu C. Advances in HBV infection and replication systems in vitro. Virol J 2021; 18(1): 105-15. doi: 10.1186/s12985-021-01580-6.
- Turton KL, Meier-Stephenson V, Badmalia MD, Coffin CS, Patel TR. Host transcription factors in hepatitis B virus RNA synthesis. Viruses 2020; 12(2) 133-42. doi: 10.3390/v120 20160.
- Xia C, Tang W, Geng P, Zhu H, Zhou W, Huang H, et al. Baicalin down-regulating hepatitis B virus transcription depends on the liver-specific HNF4α-HNF1α axis. Toxicol Appl Pharmacol 2020; 403:115131. doi: 10.1016/j.taap. 2020.115131.
- Pan Y, Ke Z, Ye H, Sun L, Ding X, Shen Y, et al. Saikosaponin C exerts anti-HBV effects by attenuating HNF1α and HNF4α expression to suppress HBV pgRNA synthesis. Inflamm Res 2019; 68(12):1025-34. doi: 10.1007/s00011-019-01284-2.
- Hatzis P, Kyrmizi I, Talianidis I. Mitogen-activated protein kinase-mediated disruption of enhancer-promoter communication inhibits hepatocyte nuclear factor 4alpha Expression. Mol Cell Biol 2006; 26(19): 7017-29. doi: 10.1128/mcb.00 297-06.
- Wang HQ, Chen FH, Wang L, Chi LQ, Wang GH. Biopharmaceutical and pharmacokinetic activities of oxymatrine determined by a sensitive UHPLC-MS/MS Method. Curr Pharm Biotechnol 2022; 23(1):148-57. doi: 10.2174/13892010 22666210118160529.
- Lan X, Zhao J, Zhang Y, Chen Y, Liu Y, Xu F. Oxymatrine exerts organ- and tissue-protective effects by regulating inflammation, oxidative stress, apoptosis, and fibrosis: From bench to bedside. Pharmacol Res 2020; 151:1041-54. doi: 10.1016/j.phrs.2019.104541.
- Wang YP, Zhao W, Xue R, Zhou ZX, Liu F, Han YX, et al. Oxymatrine inhibits hepatitis B infection with an advantage of overcoming drug-resistance. Antiviral Res 2011; 89(3): 227-31. doi: 10.1016/j.antiviral.2011.01.005.
- Lin M, Yang L, Li W, Peng Y, Zheng J. Inhibition of the replication of hepatitis B virus in vitro by oxymatrine. J Int Med Res 2009; 37(5):1411-19. doi: 10.1177/1473230009037005.
- Sang X, Wang R, Han Y, Zhang C, Shen H, Yang Z, et al. T Cell-associated immunoregulation and antiviral effect of oxymatrine in hydrodynamic injection HBV mouse model. Acta Pharm Sin B 2017; 7(3):311-8. doi: 10.1016/j.apsb. 2017.02.005.
- Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA 1987; 84(4):1005-9. doi:10.1073/pnas.84.4.1005.
- Ma ZJ, Li Q, Wang JB, Zhao YL, Zhong YW, Bai YF, et al. Combining oxymatrine or matrine with lamivudine increased its antireplication effect against the hepatitis B virus in vitro. Evid Based Complement Alternat Med 2013; 2013:186573. doi: 10.1155/2013/186573.
- Wang LJ, Yue PF, Zhao YL, Cai PL, Zhou X, Yuan HL. Preparation and study of anti-hepatitis B virus activity in vitro of oxymatrine phospholipid complex. J Chin Pharm Sci 2007; 16(2):146-52. doi: 10.1016/j.jeurceramsoc.2007.07.010.
- Wu XN, Wang GJ. Experimental studies of oxymatrine and its mechanisms of action in hepatitis B and C viral infections. Chin J Dig Dis 2004; 5(1):12-6. doi: 10.1111/j. 1443-9573.2004.00146.x.
- Yao N, Wang X. In vitro immunomodulatory activity of oxymatrine on toll-like receptor 9 signal pathway in chronic hepatitis B. Am J Chin Med 2014; 42(6):1399-10. doi: 10.1142/s0192415x 14500888.
- Gunewardena S, Huck I, Walesky C, Robarts D, Weinman S, Apte U. Progressive loss of hepatocyte nuclear factor 4 alpha activity in chronic liver diseases in humans. Hepatology 2022; 76(2):372-86. doi: 10.1002/hep.32326.
- Dai X-Q, Cai W-T, Wu X, Chen Y, Han F-M. Protocatechuic acid inhibits hepatitis b virus replication by activating ERK1/2 pathway and down-regulating HNF4α and HNF1α in vitro. Life Sci 2017; 180:68-74. doi: 10.1016/j.lfs.2017.05. 015.
- Lin J, Gu C, Shen Z, Liu Y, Wang W, Tao S, et al. Hepatocyte nuclear factor 1α downregulates HBV gene expression and replication by activating the NF-κB signaling pathway. PLoS One 2017; 12(3): 174-79. doi: 10.1371/journal.pone.0174017.
- Xie M, Guo H, Lou G, Yao J, Liu Y, Sun Y, et al. Neddylation inhibitor MLN4924 has anti-HBV activity via modulating the ERK-HNF1α-C/EBPα-HNF4α Axis. J Cell Mol Med 2021; 25(2):840-54. doi: 10.1111/jcmm.16137.
- Pan Y, Ke Z, Ye H, Sun L, Ding X, Shen Y, et al. Saikosaponin C exerts anti-HBV effects by attenuating HNF1α and HNF4α expression to suppress HBV pgRNA synthesis. Inflamm Res 2019; 68(12): 1025-34. doi: 10.1007/s00011-019-01284-2.
- He F, Chen EQ, Liu L, Zhou TY, Liu C, Cheng X, et al. Inhibition of hepatitis B virus replication by hepatocyte nuclear factor 4-alpha specific short hairpin RNA. Liver Int 2012; 32(5):742-51. doi: 10.1111/j.1478-3231.2011.02748.x.
- Zheng Y, Li J, Johnson DL, Ou JH. Regulation of hepatitis B virus replication by the ras-mitogen-activated protein kinase signaling pathway. J Virol 2003; 77(14):7707-12. doi: 10.1128/jvi.77.14.7707-7712.2003.
- Park JI. MAPK-ERK Pathway. Int J Mol Sci 2023; 24(11): 9666-69. doi.org/10.3390/ijms24119666.
- Bai L, Nong Y, Shi Y, Liu M, Yan L, Shang J, et al. Luteolin inhibits hepatitis B virus replication through extracellular signal-regulated kinase-mediated down-regulation of hepatocyte nuclear factor 4alpha expression. Mol Pharmaceutics 2016; 13(2):568-77. doi: 10.1021/acs.molpharmaceut.5b00789.